OME-NGFF
October 24th 2023
BioImage Town
 cc-by J. Moore et al.
cc-by J. Moore et al.
The venerable TIFF
(in 1986) TIFF was created as an attempt to get desktop scanner vendors of the mid-1980s to agree on a common scanned image file format, in place of a multitude of proprietary formats. wikipedia
Despite this nice standard, proprietary formats proliferated
Metadata and OME-TIFF
OME-TIFF extends TIFF to store microscopy (up to 5D) images
Metadata stored in the
Descriptionsection of the headerFollows OME-XML
Quite complete acquisition model (as of 2016)
Annotations
Issues
- Read one whole plane at a time
- Metadata model is quite rigid
- Pyramidal tiff is a bit hackish
- Only represents images
OME-NGFF
- Data is (way) bigger than memory
- Data is no longer on your computer
- We want parallelism
- Microscopy is more than images
ZARR: a hierarchical, cloud native format
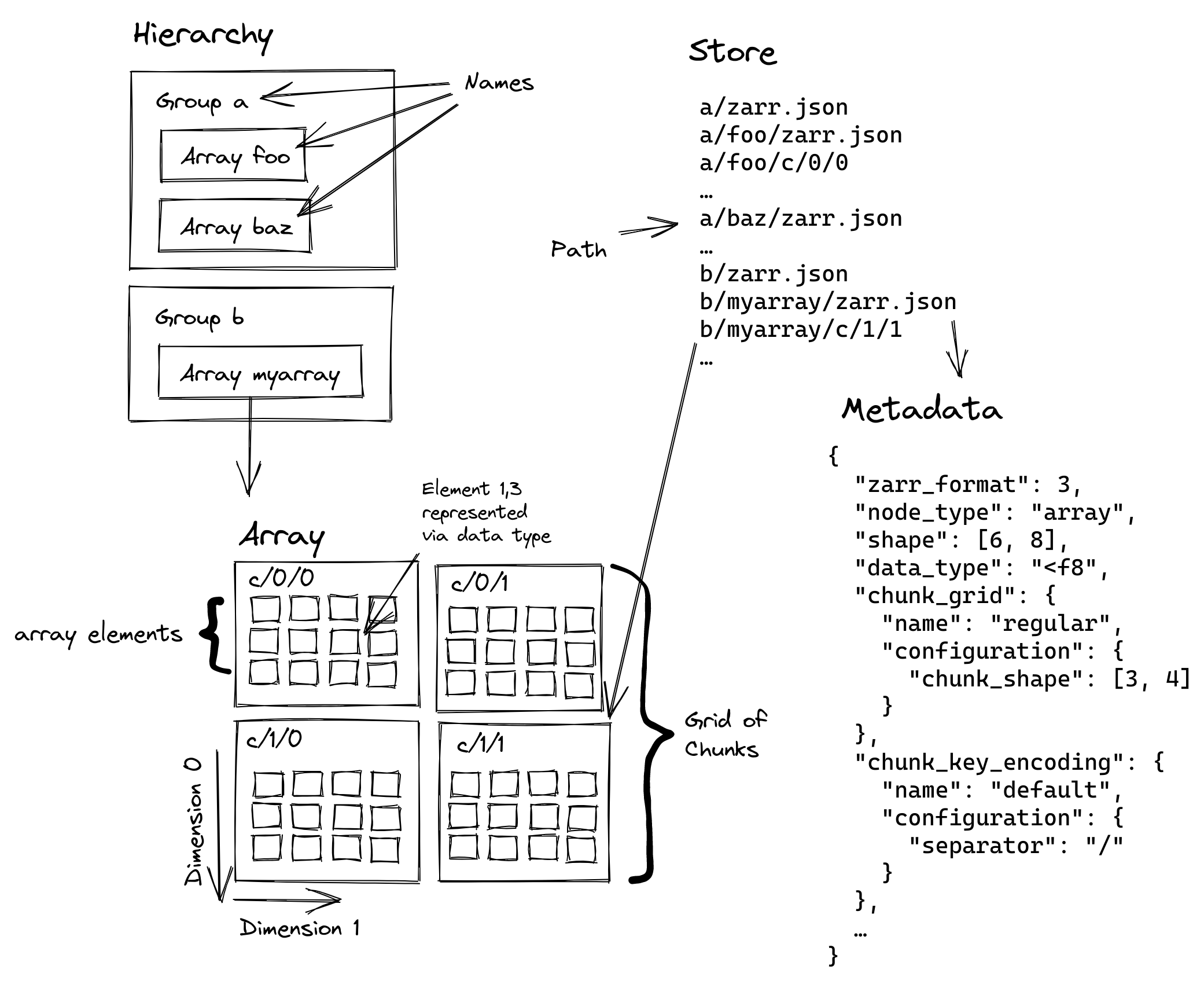
OME-ZARR
Specialisation of ZARR to describe Microscopy data
The goal is to have a FAIR data format, wihout the limitations of TIFF, adapted to non local data worklows.
Metadata
All in a single json file
1. "axes"
2. "coordinateTransformations"
3. "multiscales"4. "omero" (transitional)
5. "labels"
6. "plate"
7. "well"- Metadata can evolve more easily
- ZARR can accomodate data outside of the spec
Pyramids and Chunks
- Efficient browsing
- Efficient res-slicing
- Parallel I/O
- Parallel compute
Tools
- Python, java, js, Julia, C
- BioFormats can read zarr (see
ngff-converter) - compatible with OMERO
Napari
napari --plugin napari-ome-zarr https://uk1s3.embassy.ebi.ac.uk/idr/zarr/v0.1/4495402.zarr

Fiji and MoBie

Embeded
Online examples
Prospects
Issues
- One chunk per file -> many files (WIP: sharding)
- Still not trivial to use for non programmers
- Need a write optimized implementation
- Vendor adoption
Current work
- Specification evolution process
- How to store tables?
- How to store evolvable metadata?
Community

